rstanarmを使ってロジスティクス回帰(事前情報のlocationを変更)
library(tidyverse)
library(caret)
library(GGally)
library(ggplot2)
library(corrplot)
library(bayesplot)
theme_set(bayesplot::theme_default(base_family = "sans"))
library(rstanarm)
options(mc.cores = 1)
library(loo)
library(projpred)
SEED=14124869
#df1 少サンプル
nnn<-40
v1_vec<-round(rnorm(nnn,60,10))
v2_vec<-rbinom(nnn,1,5/10)
v3_vec<-rbinom(nnn,1,5/10)
v4_vec<-rbinom(nnn,1,5/10)
pp<-0.1+v2_vec*0.15
outcome_vec<-apply(matrix(pp),1,function(x) rbinom(1,1,x))
df1<-data.frame(v1_vec,v2_vec,v3_vec,v4_vec,outcome_vec)
#少サンプル 、無情報事前分布
df<-df1
names(df)<-c("v1","v2","v3","v4","outcome")
df[1]<-scale(df[1])
n=dim(df)[1]
p=dim(df)[2]
corrplot(cor(df[,c(1:4)]))
df$outcome<-factor(df$outcome)
x<-model.matrix(outcome ~. -1, data = df)
y<-df$outcome
(reg_formula <- formula(paste("outcome ~", paste(names(df)[1:(p-1)], collapse = " + "))))
#頻度
logistic_model <- glm(outcome ~v1 + v2 +v3 +v4, data = df, family = "binomial")
summary(logistic_model)
freq_ans<-summary(logistic_model)$coefficient #func
calc<-function(k){
t_prior <- student_t(df = c(7,7,7,7), location = c(0,k,0,0), scale = c(2.5,2.5,2.5,2.5))
t_prior_intercept <- student_t(df = 7, location = 0, scale = 2.5)
post1 <- stan_glm(reg_formula, data = df,
family = binomial(link = "logit"),
prior = t_prior, prior_intercept = t_prior_intercept, QR=TRUE,
seed = SEED, refresh = 0)
# pplot <- plot(post1, "areas", prob = 0.95, prob_outer = 1)
# pplot + geom_vline(xintercept = 0)
ans<-list(round(coef(post1), 2),
round(posterior_interval(post1, prob = 0.9), 2))
return(ans)
}
kvec<-seq(-2,2,by=0.1)
est_intercept<-est_v1<-est_v2<-est_v3<-est_v4<-c()
low_intercept<-low_v1<-low_v2<-low_v3<-low_v4<-c()
up_intercept<-up_v1<-up_v2<-up_v3<-up_v4<-c()
for(j in 1:length(kvec)){
set<-calc(kvec[j])
est_intercept[j]<-set[[1]][1]
est_v1[j]<-set[[1]][2]
est_v2[j]<-set[[1]][3]
est_v3[j]<-set[[1]][4]
est_v4[j]<-set[[1]][5]
low_intercept[j]<-set[[2]][1,1]
low_v1[j]<-set[[2]][2,1]
low_v2[j]<-set[[2]][3,1]
low_v3[j]<-set[[2]][4,1]
low_v4[j]<-set[[2]][5,1]
up_intercept[j]<-set[[2]][1,2]
up_v1[j]<-set[[2]][2,2]
up_v2[j]<-set[[2]][3,2]
up_v3[j]<-set[[2]][4,2]
up_v4[j]<-set[[2]][5,2]
}
plot(kvec,est_intercept,ylim=c(-8,0),type="o",col=2,ylab="est",xlab="prior location",main="intercept");par(new=T)
plot(kvec,low_intercept,ylim=c(-8,0),type="l",xlab="",ylab="");par(new=T)
plot(kvec,up_intercept,ylim=c(-8,0),type="l",xlab="",ylab="");
abline(h=freq_ans[1,1],col=4)
plot(kvec,est_v1,ylim=c(-4,4),type="o",col=2,ylab="est",xlab="prior location",main="v1");par(new=T)
plot(kvec,low_v1,ylim=c(-4,4),type="l",xlab="",ylab="");par(new=T)
plot(kvec,up_v1,ylim=c(-4,4),type="l",xlab="",ylab="");
abline(h=freq_ans[2,1],col=4)
plot(kvec,est_v2,ylim=c(-4,4),type="o",col=2,ylab="est",xlab="prior location",main="v2");par(new=T)
plot(kvec,low_v2,ylim=c(-4,4),type="l",xlab="",ylab="");par(new=T)
plot(kvec,up_v2,ylim=c(-4,4),type="l",xlab="",ylab="");
abline(h=freq_ans[3,1],col=4)
plot(kvec,est_v3,ylim=c(-4,4),type="o",col=2,ylab="est",xlab="prior location",main="v3");par(new=T)
plot(kvec,low_v3,ylim=c(-4,4),type="l",xlab="",ylab="");par(new=T)
plot(kvec,up_v3,ylim=c(-4,4),type="l",xlab="",ylab="");
abline(h=freq_ans[4,1],col=4)
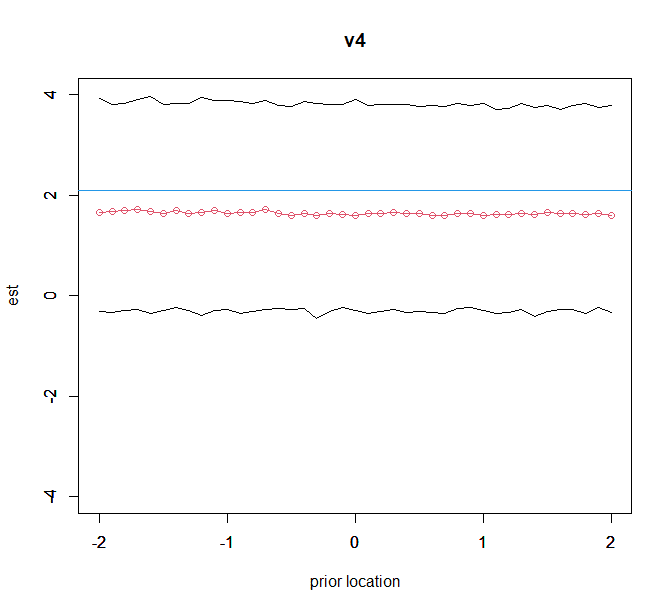
plot(kvec,est_v4,ylim=c(-4,4),type="o",col=2,ylab="est",xlab="prior location",main="v4");par(new=T)
plot(kvec,low_v4,ylim=c(-4,4),type="l",xlab="",ylab="");par(new=T)
plot(kvec,up_v4,ylim=c(-4,4),type="l",xlab="",ylab="");
abline(h=freq_ans[5,1],col=4)




追加:さらにv2のscale=0.1としたとき

この記事が気に入ったらサポートをしてみませんか?
